authors: Phil Reed, Alban Gaignard, Leyla Jael Castro
contributors: Nick Juty, Roman Baum
- Initially drafted the 20th of November 2025, as part of Bioschemas activities and ELIXIR BioHackathon Europe 2025.
- Refined at de.NBI BioHackathon Germany 2025.
- To be revised by the community.
Any questions, please contact [email protected], [email protected]
Report experiments to model Learning Paths with Schema.org ontology.
A jupyter notebook showing :
- How Schema.org can be used, especially the HowTo, HowToSection, and HowToStep types and the required properties to properly define order between steps.
- Some code to query (SPARQL and property path) the semantic annotations and retrieve for a given step the learning prerequesites.
- Some code to transform Schema.org markup into a Mermaid diagram
A sample Learning path:
LP structure:
Introduction to Galaxy and Sequence analysis [syllabusSections=M1,M2]
- Module 1: Introduction to Galaxy [itemListElement=11,12] [nextItem=M2]
- (1.1) A short introduction to Galaxy [nextItem=12]
- (1.2) Galaxy Basics for genomics [nextItem=21]
- Module 2: Basics of Genome Sequence Analysis [itemListElement=21,22,23,24]
- (2.1) Quality Control [nextItem=22]
- (2.2) Mapping [nextItem=23]
- (2.3) An Introduction to Genome Assembly [nextItem=24]
- (2.4) Chloroplast genome assembly
@prefix dct: <http://purl.org/dc/terms/> .
@prefix ex: <http://example.org/> .
@prefix schema: <https://schema.org/> .
ex:GA_learning_path a schema:Course ;
dct:conformsTo <https://bioschemas.org/profiles/LearningPath> ;
schema:courseCode "GSA101" ;
schema:description "This learning path aims to teach you the basics of Galaxy and analysis of sequencing data. " ;
schema:name "Introduction to Galaxy and Sequence analysis" ;
schema:provider ex:ExampleUniversity ;
schema:syllabusSections ex:Module_1,
ex:Module_2 .
ex:Module_1 a schema:ItemList,
schema:ListItem,
schema:Syllabus ;
dct:conformsTo <https://bioschemas.org/profiles/LearningPathModule> ;
schema:itemListElement ex:TM11,
ex:TM12 ;
schema:name "Module 1: Introduction to Galaxy" ;
schema:nextItem ex:Module_2 ;
schema:teaches "Learn how to create a workflow" .
ex:TM11 a schema:LearningResource,
schema:ListItem ;
dct:conformsTo <https://bioschemas.org/profiles/TrainingMaterial> ;
schema:description "What is Galaxy" ;
schema:name "(1.1) A short introduction to Galaxy" ;
schema:nextItem ex:TM12 ;
schema:url "https://tess.elixir-europe.org/materials/hands-on-for-a-short-introduction-to-galaxy-tutorial?lp=1%3A1" .And the complete diagram:
graph TD
N1["Module 1: Introduction to Galaxy"]
N3["(1.2) Galaxy Basics for genomics"]
N1 -- itemListElement --> N3
N1["Module 1: Introduction to Galaxy"]
N2["(1.1) A short introduction to Galaxy"]
N1 -- itemListElement --> N2
N4["Module 2: Basics of Genome Sequence Analysis"]
N8["(2.4) Chloroplast genome assembly"]
N4 -- itemListElement --> N8
N2["(1.1) A short introduction to Galaxy"]
N3["(1.2) Galaxy Basics for genomics"]
N2 -- nextItem --> N3
N1["Module 1: Introduction to Galaxy"]
N4["Module 2: Basics of Genome Sequence Analysis"]
N1 -- nextItem --> N4
N7["(2.3) An Introduction to Genome Assembly"]
N8["(2.4) Chloroplast genome assembly"]
N7 -- nextItem --> N8
N4["Module 2: Basics of Genome Sequence Analysis"]
N5["(2.1) Quality Control"]
N4 -- itemListElement --> N5
N4["Module 2: Basics of Genome Sequence Analysis"]
N6["(2.2) Mapping"]
N4 -- itemListElement --> N6
N4["Module 2: Basics of Genome Sequence Analysis"]
N7["(2.3) An Introduction to Genome Assembly"]
N4 -- itemListElement --> N7
N6["(2.2) Mapping"]
N7["(2.3) An Introduction to Genome Assembly"]
N6 -- nextItem --> N7
N3["(1.2) Galaxy Basics for genomics"]
N5["(2.1) Quality Control"]
N3 -- nextItem --> N5
N5["(2.1) Quality Control"]
N6["(2.2) Mapping"]
N5 -- nextItem --> N6
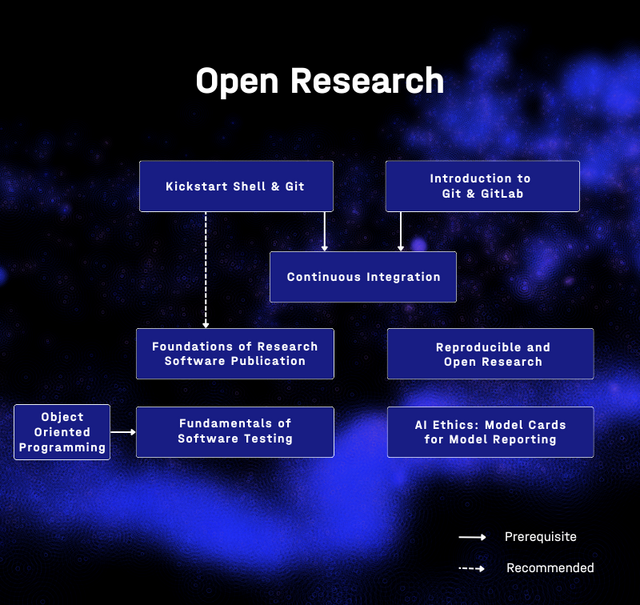
This is a module of a learning path graph where each item is a course instance (event), not a training material. They provide a linear reduction which we will model. This reduction supports the prerequisites in a simplified way, and ignores courses from other modules (Open Oriented Programming).
LP structure:
Helmholtz Data Science Course Portfolio [syllabusSections=M1]
- Module: Open Research [itemListElement=1,2,3,4,5,6,7]
- (1) Kickstart Shell & Git [nextItem=2]
- 22-23 September 2025
- (2) Introduction to Git & GitLab [nextItem=3]
- 15-16 September 2025
- 24-25 November 2025
- (3) Foundations of Research Software Publication [nextItem=4]
- 6-7 November 2025
- (4) Continuous Integration [nextItem=5]
- 28-29 October 2025
- 16-17 December 2025
- (5) Fundamentals of Software Testing [nextItem=6]
- No instances yet
- (6) Reproducible and Open Research [nextItem=7]
- 19 November 2025
- (7) AI Ethics: Model Cards for Model Reporting
- 17 October 2025
- (1) Kickstart Shell & Git [nextItem=2]
Each course may have multiple course instances.
hz:HZ_learning_path a schema:Course ;
dct:conformsTo <https://bioschemas.org/profiles/LearningPath> ;
schema:courseCode "data-science-course-portfolio" ;
schema:description "The platforms of the Information & Data Science Framework have developed a coordinated course offer that covers a wide array of topics." ;
schema:name "Helmholtz Data Science Course Portfolio" ;
schema:provider hz:Helmholtz ;
schema:syllabusSections hz:Module_1 .
hz:Module_1 a schema:Course,
schema:ItemList,
schema:ListItem,
schema:Syllabus ;
dct:conformsTo <https://bioschemas.org/profiles/LearningPathModule> ;
schema:description "Unlock the potential of open science through courses on collaborative research practices and project management, open-source tools and software, and publishing ethically sound, reproducible, and transparent results." ;
schema:itemListElement hz:TM1,
hz:TM2,
hz:TM3,
hz:TM4,
hz:TM5,
hz:TM6,
hz:TM7 ;
schema:name "Open Research" ;
schema:teaches "Learn how to support open science" .
hz:TM1 a schema:Course,
schema:ListItem ;
dct:conformsTo <https://bioschemas.org/profiles/Course> ;
schema:description "Boost your research efficiency with this workshop on Bash scripting and Git version control. Learn to automate tasks with custom scripts and track your work with Git" ;
schema:hasCourseInstance hz:TM1a ;
schema:name "(1) Kickstart Shell & Git" ;
schema:nextItem hz:TM2 ;
schema:url "https://www.helmholtz-hida.de/en/lernen-vernetzen/data-science-course-portfolio/kickstart-shell-git/" .
hz:TM1a a schema:CourseInstance ;
dct:conformsTo <https://bioschemas.org/profiles/CourseInstance> ;
schema:courseMode "online" ;
schema:endDate "2025-09-23T17:00:00" ;
schema:startDate "2025-09-22T09:00:00" .And the complete diagram:
graph TD
N2["(1) Kickstart Shell & Git"]
N3["(2) Introduction to Git & GitLab"]
N2 -- nextItem --> N3
N1["Open Research"]
N8["(7) AI Ethics: Model Cards for Model Reporting"]
N1 -- itemListElement --> N8
N5["(4) Continuous Integration"]
N6["(5) Fundamentals of Software Testing"]
N5 -- nextItem --> N6
N3["(2) Introduction to Git & GitLab"]
N4["(3) Foundations of Research Software Publication"]
N3 -- nextItem --> N4
N1["Open Research"]
N5["(4) Continuous Integration"]
N1 -- itemListElement --> N5
N1["Open Research"]
N3["(2) Introduction to Git & GitLab"]
N1 -- itemListElement --> N3
N1["Open Research"]
N7["(6) Reproducible and Open Research"]
N1 -- itemListElement --> N7
N4["(3) Foundations of Research Software Publication"]
N5["(4) Continuous Integration"]
N4 -- nextItem --> N5
N1["Open Research"]
N2["(1) Kickstart Shell & Git"]
N1 -- itemListElement --> N2
N6["(5) Fundamentals of Software Testing"]
N7["(6) Reproducible and Open Research"]
N6 -- nextItem --> N7
N1["Open Research"]
N4["(3) Foundations of Research Software Publication"]
N1 -- itemListElement --> N4
N1["Open Research"]
N6["(5) Fundamentals of Software Testing"]
N1 -- itemListElement --> N6
N7["(6) Reproducible and Open Research"]
N8["(7) AI Ethics: Model Cards for Model Reporting"]
N7 -- nextItem --> N8
We propose two new Bioschemas profiles and a small change to one Bioschemas profile:
LearningPath: inherits fromCourseLearningPathModule: inherits fromCourseandSyllabusandListItemandItemListTrainingMaterial: inherits fromLearningResourceandListItem
A LearningPath has zero or more LearningPathModule. A LearningPathModule has zero or more LearningResource. These relationships are (ordered) lists or steps, using the ItemList and Syllabus Schema.org types.
Class diagram, where 🔺 is Schema.org type, 🟩 is Bioschemas profile, 🔵 is new profile:
classDiagram
direction TB
class Event["Event🔺"] {
}
class CourseInstance["CourseInstance🔺🟩"] {
}
class Course["Course🔺🟩"] {
syllabusSections
}
class new_LearningPath["new:LearningPath🔵"] {
Syllabus[] syllabusSections
}
class ListItem["ListItem🔺"] {
nextItem
}
class Syllabus["Syllabus🔺"] {
}
class new_LearningPathModule["new:LearningPathModule🔵"] {
ListItem[] itemListElement
LearningPathTopic nextItem
}
class LearningResource["LearningResource🔺"] {
}
class bio_TrainingMaterial["bio:TrainingMaterial🟩"] {
}
Course <|-- new_LearningPath
Course <|-- new_LearningPathModule
Syllabus <|-- new_LearningPathModule
ListItem <|-- new_LearningPathModule
LearningResource <|-- Course
LearningResource <|-- bio_TrainingMaterial
LearningResource <|-- Syllabus
Event <|-- CourseInstance
More detailed class diagram, including the distinction where there is a Schema.org type with the same name as a Bioschemas profile:
classDiagram
direction TB
class CourseInstance["CourseInstance🔺"] {
}
class bio_CourseInstance["bio:CourseInstance🟩"] {
}
class bio_Course["bio:Course🟩"] {
}
class Course["Course🔺"] {
syllabusSections
}
class new_LearningPath["new:LearningPath🔵"] {
Syllabus[] syllabusSections
}
class ListItem["ListItem🔺"] {
nextItem
}
class Syllabus["Syllabus🔺"] {
}
class new_LearningPathModule["new:LearningPathModule🔵"] {
ListItem[] itemListElement
LearningPathTopic nextItem
}
class LearningResource["LearningResource🔺"] {
}
class bio_TrainingMaterial["bio:TrainingMaterial🟩"] {
}
class CreativeWork["CreativeWork🔺"] {
}
class Event["Event🔺"] {
}
Course <|-- bio_Course
Course <|-- new_LearningPath
Course <|-- new_LearningPathModule
Syllabus <|-- new_LearningPathModule
ListItem <|-- new_LearningPathModule
CreativeWork <|-- LearningResource
CreativeWork <|-- Course
LearningResource <|-- Course
LearningResource <|-- Syllabus
Event <|-- CourseInstance
CourseInstance <|-- bio_CourseInstance
LearningResource <|-- bio_TrainingMaterial